Tracking genetic history of a population
A Genetic Detective Story
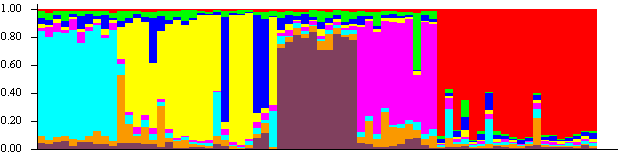
My group and I completed a project in which we and another team simulated the genetics of two populations, one inspired by the Halo universe and the other inspired by the Star Wars universe. We then switched to analyze the opposing groups’ data, trying to discover historical events and population changes without knowing what they actually modeled.
We used a program called SLiM to simulate the genetics of our populations. SLiM is a free, open-source software package that can be used to simulate the evolution of populations.
We started by creating a population of 10,000 individuals for each of our two groups. We then ran the simulation for 100,000 generations. During this time, we allowed the populations to evolve through mutation, recombination, and migration.
Once the simulation was complete, we analyzed the data from each population using a variety of statistical methods. We looked at things like the nucleotide diversity (Pi), the effective population size (Ne), and the CLR test.
The Pi statistic is a measure of the amount of genetic variation in a population. A high Pi value indicates that there is a lot of genetic variation, while a low Pi value indicates that there is little genetic variation.
The effective population size is a measure of the number of individuals that are actually contributing genes to the next generation. A large Ne value indicates that there is a lot of genetic mixing, while a small Ne value indicates that there is less genetic mixing.
The CLR test is a statistical test that can be used to identify regions of the genome that have been subject to positive or negative selection.
By analyzing the data from our two populations, we were able to learn a lot about their history. We found that the Halo population had experienced a number of selective sweeps, which are events where a beneficial mutation becomes fixed in the population. We also found that the Star Wars population had experienced a lot of migration, which is the movement of individuals between populations.
We were able to use this information to reconstruct the history of our two populations. We believe that the Halo population was originally a single population, but it split into two populations at some point in the past. One of these populations eventually became the Covenant, while the other population became the humans.
We believe that the Star Wars population was also originally a single population, but it experienced a lot of migration over time. This migration led to the formation of the different species that we see in the Star Wars universe.
We were very excited to be able to use genetic data to learn about the history of two fictional universes; the same technology can also be used to look back at real population data to find historical events in their past.
[text]